Research
Cell Villages
Cell villages are the core of our research. We think they’re awesome and we think you will too.
The Wells Lab is driven to uncover the mysteries surrounding the developing human brain. We aim to answer major questions like:
- How do variants in the human genome influence neurodevelopmental processes?
- What are the mechanisms underlying neurodevelopmental disorders, and how can they be corrected?
- What technologies can we devise to speed up the pace of discovery?
To address these challenges, we use 2D and 3D in vitro human stem cell-derived neural cell models, CRISPR-Cas9 gene editing, and a revolutionary experimental platform known as a cell village. This system involves the pooling of stem cell lines and their derivatives from multiple human donors into the same culture flask, thus capturing genetic, molecular, and phenotypic heterogeneity in a shared in vitro environment. This approach can be used to perform unbiased, whole-genome and transcriptome-wide screens of various cellular phenotypes, while minimizing the technical variation that often plagues multi-line comparative investigations and cuts down on the financial costs of culturing hundreds of cell lines independently.
The village platform is enabled by two bioinformatic pipelines:
- Census-seq, which can provide insight into phenotypic differences across donors; and
- Dropulation, which can define the gene expression profile and donor-of-origin of each cell in the village.
From here, we can deploy a series of statistical tools such as expression quantitative trait loci detection, group-wise differential gene expression analysis, and genome- or transcriptome-wide association tests to describe the relationships among alleles, gene expression, and cellular phenotypes in a high-throughput manner.
Here are three ongoing lines of investigation that we are pursuing in the Wells Lab:
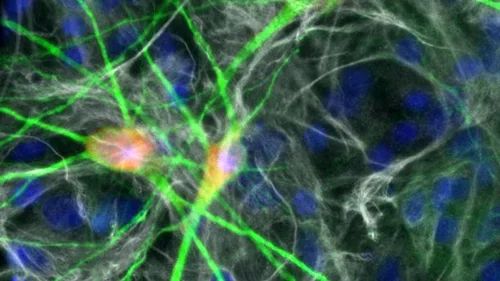
16p11.2 Microdeletion Syndrome
Autism spectrum disorders (ASD) affect 1 in 54 children in the United States, and the deletion or duplication of the 16p11.2 genomic region accounts for ~1% of these cases. The mechanisms underlying these symptoms are unknown, and no effective treatments are available for this disease.
We are deploying villages of 16p11.2 patient neural cells, large-scale CRISPR screens, and 3D cerebral organoids to answer the following questions:
- Which of the 29 genes in this region are contributing to disease?
- How do patient and control cells differentially respond to major signaling events, like sonic hedgehog or WNT activation?
- Are there dysfunctional molecular and cellular mechanisms shared with other genetic risk factors of ASD?
This work has the potential to identify key genetic, molecular, and cellular targets for future therapeutic interventions.
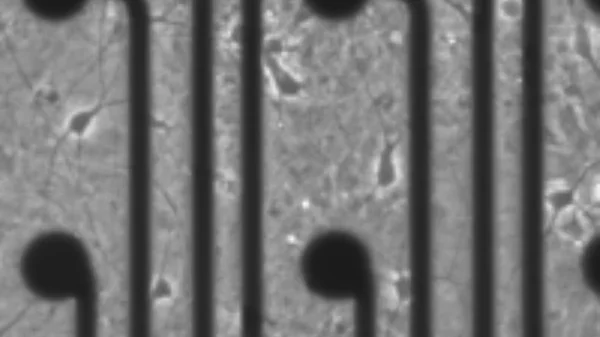
Contributors to Phenotype Diversity
Our species is characterized by an immense diversity in neurological and psychological traits. While the origins of these differences have been debated for millenia, we are starting to more fully appreciate that much of this variation is innate and could be driven by genetic and molecular factors that alter neurodevelopmental trajectories.
Using in vitro cell villages composed of donors from many different ancestral backgrounds, we are working to pinpoint the genetic variants and molecular profiles associated with neural phenotypic heterogeneity, including proliferation, differentiation, migration, and synaptic formation.
This work has the potential to uncover novel contributors to the wide array of differential human neurodevelopmental outcomes under both neurotypical and disease contexts.
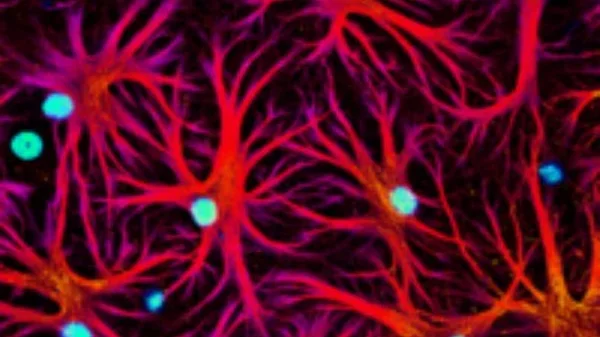
Cell Villages 2.0
The cell village platform has the potential to revolutionize the life sciences, and in theory can be applied to study any mechanism in any cell type from any human population.
Villages can be studied under static conditions in which no perturbations to the system are made, or under dynamic conditions in which the cells are subjected to various challenges, such as hypoxia, electrical stimulation, or signaling pathway activation. Villages could also be used as part of drug screens to determine how genetics influence the efficacy of candidate or FDA-approved pharmaceuticals.
To make this a reality, we are developing novel cell induction, phenotyping, and bioinformatic approaches to expand the number of cell types and phenotypes that can be assayed in a village.
This work has the potential to expand the utility of the village platform beyond the scope of the human brain.